Using the wolfram engine
Script based segmentation without mathematica
Everyting in QMRITools can also run from the command line using the Wolfram engine. For segmentation there are dedicated scripts that install and run the segmentation software. Below is a short manual on how to use these tools. The scripts and so example data can be found here.
Wolfram engine and script
- Install the free Wolfram Engine as described here:
https://www.wolfram.com/engine/ https://support.wolfram.com/45743
- Install WolframScript as described here
https://reference.wolfram.com/language/workflow/InstallWolframScript.html
QMRITools
Install the latest version of QMRITools for the worlfram enging using the Install_QMRITools.wls WolframScript.
wolframscript -f "path to file\Install_QMRITools.wls"
To install an other version of the QMRITools from the gihub release pages use
wolframscript -f "path to file\Install_QMRITools.wls" https://github.com/mfroeling/QMRITools/releases/download/4.x.x/QMRITools-4.x.x.paclet
For the situation where you run the scrip from its own folder with the test folder also there this will be script
wolframscript -f "Install_QMRITools.wls" https://github.com/mfroeling/QMRITools/releases/download/4.x.x/QMRITools-4.x.x.paclet
Segmentation script
Run the segmentation script (networks are currelty for out-phase dixon data):
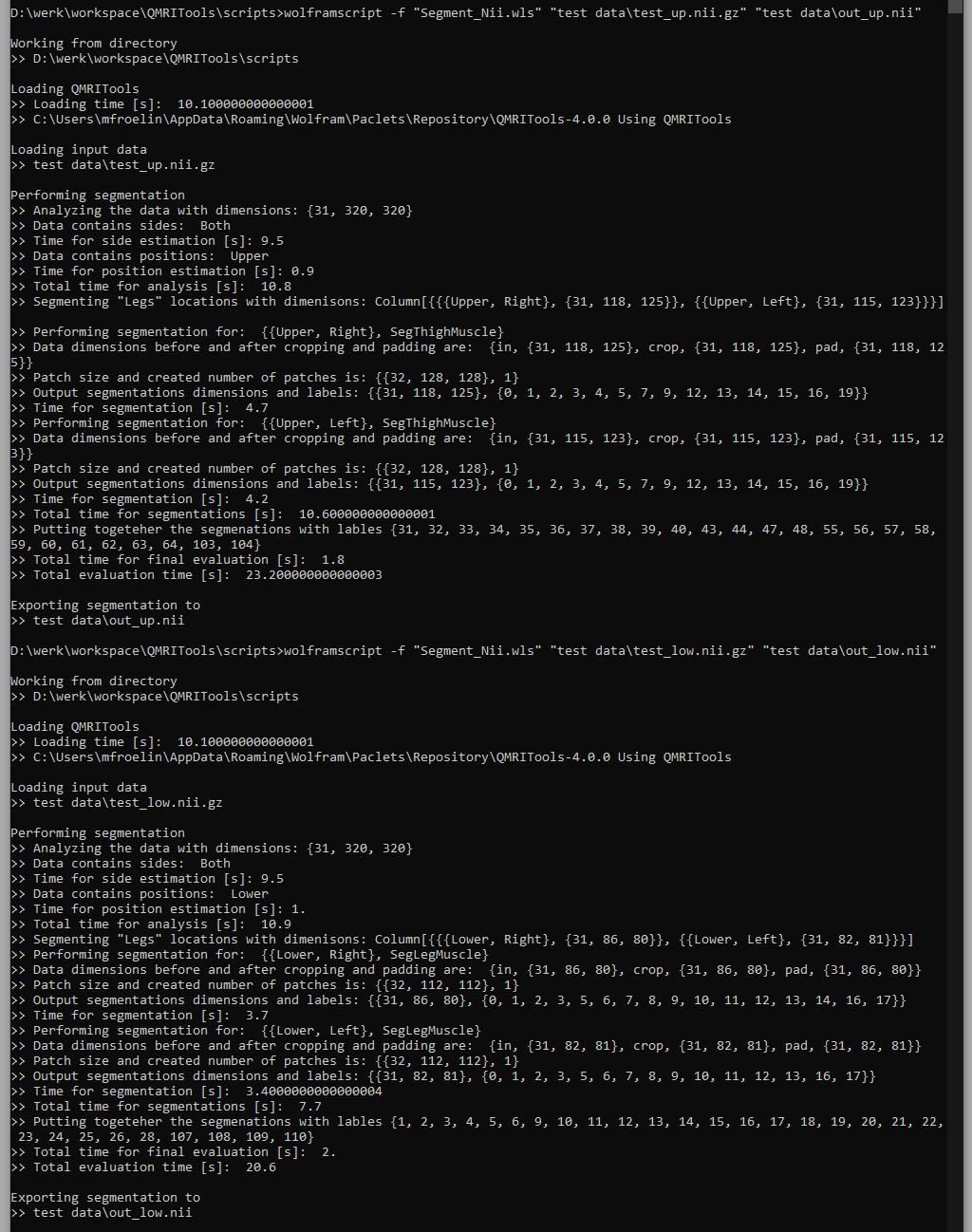
wolframscript -f "path to file\Segment_Nii.wls" "file to be segmented.nii.gz" "output file.nii"
It does not matter what part of the leg it is and if one or two legs are in the field of view. For the situation where you run the scrip from its own folder with the test folder also there this will be script.
wolframscript -f "Segment_Nii.wls" "test data\test_up.nii.gz" "test data\out_up.nii"
wolframscript -f "Segment_Nii.wls" "test data\test_low.nii.gz" "test data\out_low.nii"
By default the CPU is used, if you want to switch to GPU open the script in any text editor and change “CPU” for “GPU”.